Characterization and gene expression of heat shock protein 90 in marine crab Charybdis japonica following bisphenol A and 4-nonylphenol exposures
Article information
Abstract
Objectives
Heat shock protein 90 (HSP90) is a highly conserved molecular chaperone important in the maturation of a broad spectrum of protein. In this study, an HSP90 gene was isolated from Asian paddle crab, Charybdis japonica, as a bio-indicator to monitor the marine ecosystem.
Methods
This work reports the responses of C. japonica HSP90 mRNA expression to cellular stress by endocrine disrupting chemicals (EDCs), such as bisphenol A (BPA) and 4-nonylphenol (NP) using real-time. reverse transcription polymerase chain reaction.
Results
The deduced amino acid sequence of HSP90 from C. japonica shared a high degree of homology with their homologues in other species. In a phylogenetic analysis, C. japonica HSP90 is evolutionally related with an ortholog of the other crustacean species. The expression of HSP90 gene was almost distributed in all the examined tissues of the C. japonica crab but expression levels varied among the different body parts of the crabs. We examined HSP90 mRNA expression pattern in C. japonica crabs exposed to EDCs for various exposure times. The expression of HSP90 transcripts was significantly increased in C. japonica crabs exposed to BPA and NP at different concentrations for 12, 24, 48 and 96 hours. The mRNA expression of HSP90 gene was significantly induced in a concentration- and time-dependent manner after BPA or NP exposures for 96 hours.
Conclusions
Taken together, expression analysis of Asian paddle crab HSP90 gene provided useful molecular information about crab responses in stress conditions and potential ways to monitor the EDCs stressors in marine environments.
Introduction
Cells generally respond to stress by changing gene expression and up-regulating the production of a group of highly conserved proteins known as the heat shock proteins (HSPs) [1]. Heat shock protein 90 (HSP90) are important molecular chaperones that contribute to the proper folding of cellular proteins. They associate with steroid hormone receptors and maintain them in a non-functional state until hormone binding and interact with cellular signaling proteins [2,3]. HSP90 genes have been isolated from various crustaceans including lobsters [4], shrimps [5,6] and crabs [3,7]. HSP90 mediates estrogen signal transduction, which regulates the synthesis of vitellogenin [5]. It was demonstrated in lobster Homarus americanus that osmotic stress significantly increased the levels of HSP90 mRNA in the abdominal muscle [8]. HSP90 may play an important role in salinity tolerance in juvenile mitten crabs, Eriocheir sinensis [1]. HSP90 may be involved in the defense system or the stress response of the blue crab, Callinectes sapidus. The injection of lipopolysaccharides caused the greatest increment of C. sapidus HSP90 [9]. In aquatic animals, the induction of HSP90 genes has been widely reported in response to thermal shock [6,10], osmotic stress [11], hypoxia [3], an exposure of heavy metals [12,13] and virus infections [14]. Additionally, the HSP90s gene expression level had even been characterized as a sensitive marker in the aquaculture industry to examine the farming condition and optimize the rearing density of fish [15]. Recent studies reported that HSP90 is involved in developmental and physiological regulation processes of crustaceans [5,16,17]. However, little is known about endocrine-related functions of crab HSP90 genes on brachyuran.
Endocrine-disrupting chemicals (EDCs) alter cellular and organ system homeostasis by interfering with the body's normal physiologic processes [18,19,20]. Industrial and municipal effluents are important sources of EDCs discharged into the aquatic environment. Bisphenol A (BPA) and 4-nonylphenol (NP) are also known EDCs ubiquitous in the aquatic environment [20]. The distribution of these EDCs has been found in coastal species as well as in the marine environment [21,22,23]. BPA is produced high volume from the manufactures used in a wide variety of consumer products [21,24]. NP is widely used in the production of nonylphenol ethoxylates, which are used in a wide variety of industrial applications and consumer products [25]. They are absorbed into marine sediment and are ubiquitously contaminated in wildlife animals and in humans and can interfere with endocrine systems [26].
The Asian paddle crab (Charybdis japonica), a marine crab, is generally found in mud flat of estuaries regions in Korea, Japan and Malaysia [27]. The crab is a potential bio-indicator species to monitor the marine sediment as well as a commercially important species living along coastal areas in Korea [28]. However, the research information of stress response in this species is still limited [28,29]. Therefore, in the present study, an HSP90 gene from C. japonica crab was isolated for the first time by cDNA library screening method and its expression analysis in response to both BPA and NP exposure were performed.
Materials and Methods
Animal Preparation
The C. japonica (Crustacea: Decapoda: Portunidae) crabs (avaverage 6-8 cm and 250 g) were collected from Aquatic Market (Yeosu, Korea). Ten marine crabs were placed in aerated glass aquaria filled with natural seawater and fed for a week. Acclimation condition for crabs were a 20±1℃ temperature, 25‰ salinity, and an 12:12 hours light-dark cycle for at least one week. Mature health crabs were only selected, and the aquaria water was constantly aerated and changed at every day.
Chemical Endocrine-disrupting Chemicals Treatments
Chemical BPA and NP were obtained from Sigma-Aldrich (St. Louis, MO, USA) and diluted with 99% acetone for stock solutions at room temperature. Using solutions were made by diluting the stock solution in sea water.
During treatment experiment, marine crabs were kept in glass tank with aerated sea water in the same acclimatization conditions. For chemical treatments, nominal concentrations of chemicals were maintained by adding fresh prepared BPA or NP. Solvent control was to adding the actual test concentration of <0.5% acetone.
Ten crabs were treated with each concentration of BPA or NP (0.05, 0.5 or 1 mg/L) for sub-lethal exposure experiments. Treatment concentrations were based on the previous studies of the acute toxicity test [28]. Exposure time was from 12 hours to 96 hours and all experiments were measured in triplicate.
Characterized Heat Shock Protein 90 Gene in C. japonica Crabs
The specific primers of C. japonica HSP90 gene designed using consensus comparison of Brachyura sequences. Multiple sequence alignments were conducted using ClustalW in Mega 4.0. Specific primers of C. japonica HSP90 were designed from orthologue sequences of Chinese mitten crab (Eriocheir sinensis, GenBank accession No. ADE60732), swimming crab (Portunus trituberculatus, GenBank accession No. ACQ90225) and green mud crab (Scylla paramamosain GenBank accession No. ACN54679). Information of primer sequences was shown in Table 1. The size of polymerase chain reaction (PCR) products were 442 bp for the HSP90 gene and 233 bp for the beta-actin gene. The condition of PCR mixture with a total volume of 50 µL contained 1×Taq polymerase buffer, 200 µM dNTP, 5 units of Taq polymerase, primers at concentrations of 10 µM and 0.5 M betaine. The PCR conditions were finally optimized as this protocol: one cycle of 94℃ for 5 minutes, 40 cycles of 94℃ for 30 seconds, 55℃ for 40 seconds, 72℃ for 1 minute and one cycle of 72℃ for 7 minutes. The amplified PCR products were cloned in the T-vector (Invitrogen, Inc., Carlsbad, CA, USA) and sequencing with an ABI 3700 genetic analyzer (Applied Biosystems, Foster City, CA, USA).
RNA Extraction and cDNA Preparation
Total RNA and cDNA from crabs were prepared using Trizol reagent (Invitrogen) and the SuperScript III reverse transcriptase kit (Invitrogen) following with the manufacturer's protocols [30]. Total RNA was extracted from various tissues in crab and hepatopancreas of C. japonica crab for EDCs exposure experiments. RNA samples were treated with DNase I to remove contaminated genomic DNA for further experiments. HSP90 gene expression in different tissues analyzed from total RNA of control C. japonica crabs. The RNA quantity was measured by a UV-visible spectrometer (SmartSpec 3000; Bio-Rad, Hercules, CA, USA). Single strand cDNA was synthesized from 3 µg RNA by using the SuperScript III reverse transcriptase kit.
Real-time Reverse Transcription Polymerase Chain Reaction of Heat Shock Protein 90 mRNA
The temporal expression of HSP90 in Asian paddle crab tissues including ovary, gill, heart, stomach, hepatopancreas, eyestalk and muscle was examined by real-time reverse transcription polymerase chain reaction (RT-PCR). The prepared cDNA were then used as PCR templates using HSP90 specific primers (Table 1). The beta-actin was amplified as an internal control [31].
Quantitative RT-PCR amplifications and measurements were carried out using an AB 7300 Real-Time PCR System (Applied Biosystems). The PCR for HSP90 was performed with 25 µL of a mixture containing 1 µL of cDNA template, 0.2 µM of each primer and 1×SYBR® Premix Ex Taq (Takara, Kyoto, Japan). For HSP90, this mixture was subjected to 33 cycles of amplification (denaturation for 30s at 94℃, annealing for 40s at 55℃ and extension for 50s at 72℃). The quality of the amplification was identified using an AB 7300 Real-Time PCR instruments (Applied Biosystems) to carry out dissociation curve analysis. The cycle threshold (Ct) was identified using the AB 7300 System SDS program. The expression levels were normalized against the beta-actin expression using the Ct method. Each experiment was calculated in triplicate.
Phylogenetic Analysis in Various Species
To analyze the phylogenetic relationship of C. japonica HSP90 gene, we aligned various species HSP90 genes at the amino acid levels characterized in our study using Clustal X version 1.8. Next, the display of multiple alignments was carried out using the GeneDoc version 2.6.001. Phylogenetic tree of C. japonica HSP90 protein were made using the neighbor-joining method of MEGA 4.0.
Network Analysis of Heat Shock Protein 90 Sequences
For network analysis of C. japonica HSP90 genes, we aligned various species HSP90 genes at the amino acid levels using Clustal X version 1.8. Next, the display of multiple alignments were carried out using the GeneDoc version 2.6.001. The HSP90 sequence relationships among species were analyzed using the NETWORK program (version 4.610). The network analysis consisted of at least three replicates.
Statistical Analysis
All data were presented as mean±standard deviation. Expression levels of HSP90 gene were normalized against the level of beta-actin using standard curves. Data of the fold change of HSP90 mRNA were logarithmically transformed before subjected to a one-way analysis of variance (ANOVA) using SPSS version 12.0 (SPSS Inc., Chicago, IL, USA). When overall differences were significant, Tukey's test was conducted to compare the means between individual treatments. The significant results were showed at p<0.05.
Results
Heat Shock Protein 90 Gene Characterization in C. japonica Crab
The partial cDNA of the C. japonica HSP90 gene was characterized from PCR product amplified using primers designed based on sequences retrieved from GenBank. We obtained a 315 bp nucleotide sequence of C. japonica HSP90 (GenBank accession No. KJ599647 in this study) that shared 94% identity with other crab HSP90 sequences (Figure 1). The amino acid sequence of C. japonica HSP90 is over 97% homologous with the corresponding gene of other crabs (Figure 1A). Sequence homology of HSP90 among Brachyura has been reported at deduced amino acids. Furthermore, the region of deduced amino acids in HSP90 was 97%, 96%, 95% and 93% similar to that of Homarus americanus (American lobster), Penaeus monodon (black tiger shrimp), Litopenaeus vannamei (Pacific white shrimp) and Metapenaeus ensis (greasyback shrimp), respectively (Figure 1B). In the phylogenetic tree, C. japonica HSP90 clustered with other crabs, lobster, oyster and shrimps, while human HSP90 formed another cluster with cattle, goat, platypus, fly and other related species (Figure 1B).
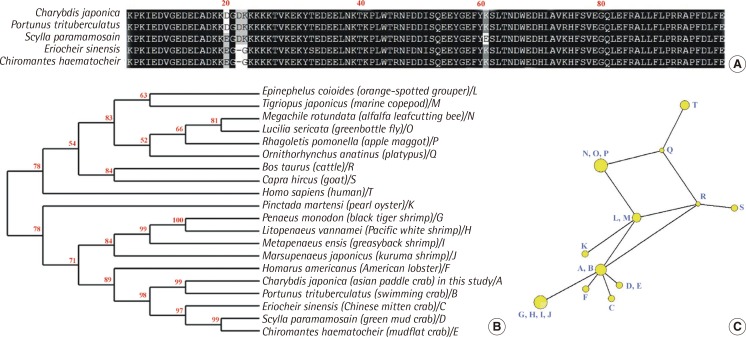
Genome analysis of heat shock protein 90 (HSP90) gene in the Asian paddle crab, Charybdis japonica. (A) Multiple sequence alignment of the deduced C. japonica HSP90 with the sequences of crabs. (B) Phylogenetic tree of the HSP90 gene constructed via neighbor-joining analysis (bootstrap value 1,000). Numbers at the nodes are the percentage bootstrap values. The GenBank accession numbers for HSP90s are: C. japonica HSP90 (KJ599647); P. trituberculatus (ACQ90225); E. sinensis (ADE60732); S. paramamosain (ACN54679); C. haematocheir (AAS19788); H. americanus (AAW34065); P. monodon (ACO83357); L. vannamei (ADU03767); M. ensis (ABR66910); M. japonicas (BAJ78983); P. martensi (AGS44978); E. coioides (AEA39713); T. japonicas (ACA03524); M. rotundata (AAS57866); L. sericata (BAD12048); R. pomonella (ABN55912); O. anatinus (XP_001518700); B. taurus (CAC84136); C. hircus (AAN40799); H. sapiens (AAH07327). (C) Network tree of the HSP90 genes among 20 species constructed by NETWORK analysis. The yellow nodes show clustering indication of species with a phylogenetic close relationship. Amino acid sequences were aligned using Clustal X version 1.8. The HSP90 sequences retrieved from the GenBank are presented in (B).
To examine the protein-protein relationship of HSP90 genes, we used a NETWORK analysis, which integrates the species-species interaction data based on HSP90 genes and provides a network construction by using polymorphic variation of HSP90 protein sequences. Figure 1C shows the evolutional network tree among twenty HSP90 sequences. The largest one of 12 nodes (yellow circles) contained related shrimps (G, H, I, J). On the network tree, C. japonica HSP90 clustered with other crabs and lobster (A, B, C, D, E and F). The C. japonica HSP90 gene has an evolutionally close relationship with a lobster, as indicated by the short branch length among nodes (Figure 1C).
Tissues mRNA Expression of C. japonica Heat Shock Protein 90
Expression of C. japonica HSP90 mRNA in various tissues was analyzed by real-time RT-PCR (Figure 2). The level of C. japonica HSP90 transcript was differently observed in studied tissues. Relatively high expressions of C. japonica HSP90 mRNA were evident in hepatopancreas, gill and ovary. The C. japonica HSP90 gene were somewhat expressed in muscle if compared with eyestalk. There was no difference in C. japonica HSP90 gene expression between crab eyestalk, heart and stomach (p>0.05).
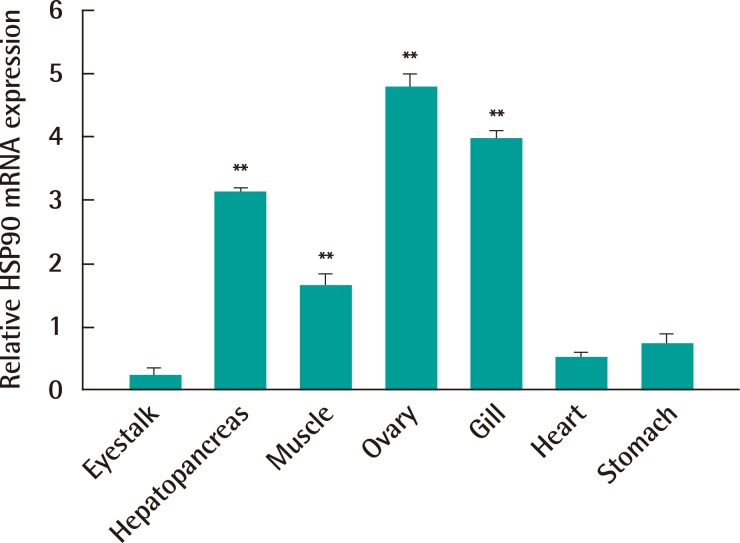
Expression of heat shock protein 90 (HSP90) gene in various tissues of a Charybdis japonica crab. Ovary, gill, heart, stomach, hepatopancreas, eyestalk and muscles were collected from 10 crabs. beta-actin gene was used as an internal control to calibrate the cDNA template for all samples. mRNA expression is shown relative to beta-actin expression after normalization. The experiment was conducted in triplicate and the data are expressed as mean±deviation. Significant differences across control (expression level of HSP90 gene in eyestalk tissue) are indicated with **p<0.01.
Responses of C. japonica Heat Shock Protein 90 mRNA to Endocrine-disrupting Chemicals Treatments
We investigated the responses of the HSP90 mRNA expression in C. japonica crab by EDC toxicity, such as BPA or NP, for from 12 to 96 hours. Figure 3 shows the expression of the HSP90 transcript in C. japonica crab in response to NP exposure with three concentrations for 12, 24, 48 and 96 hours. After the exposure to NP for 12 hours, the expression of HSP90 mRNA significantly increased in C. japonica crab exposed to only 0.5 mg/L NP of three concentrations. The HSP90 gene expression significantly increased in C. japonica crab exposed to relative high concentrations; 0.5 and 1 mg/L NP for 48 hours. An induced mRNA expression of C. japonica HSP90 was observed in NP exposures of all concentrations for 24 and 96 hours (Figure 3). The expression of the HSP90 gene was highly changed in crabs after 1 mg/L NP exposure for 96 hours (p<0.05). The significant expression of C. japonica HSP90 gene after NP exposure for 96 hours clearly observed as a dose-dependent manner.
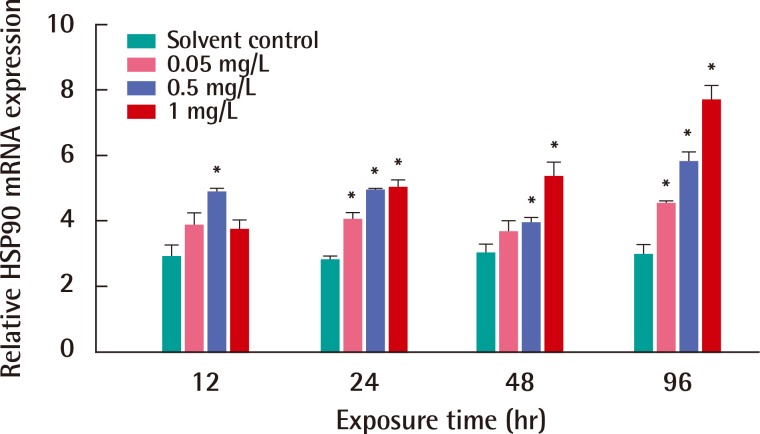
Expressions of Charybdis japonica heat shock protein 90 (HSP90) mRNA after exposure to various concentrations of 4-nonylphenol for 12, 24, 48 and 96 hr. mRNA expression is shown relative to beta-actin expression after normalization. The experiment was conducted in triplicate and the data are expressed as mean±deviation. Differences between the treatment and solvent control samples were considered to be statistically significant when *p<0.05.
After exposures of BPA, mRNA expression of HSP90 gene was observed in C. japonica crab for various exposure periods (Figure 4). HSP90 gene expression was significantly increased in C. japonica exposed to all concentrations of BPA for 12, 24, 48 and 96 hours (p<0.05). The highest expression of the HSP90 gene was observed after an exposure to the relatively high BPA concentration of 1 mg/L (p<0.01) for 96 hours. In BPA toxicity, HSP90 mRNA expression induced in C. japonica crab for the exposure times of 12 and 96 hours as a dose-dependent manner.
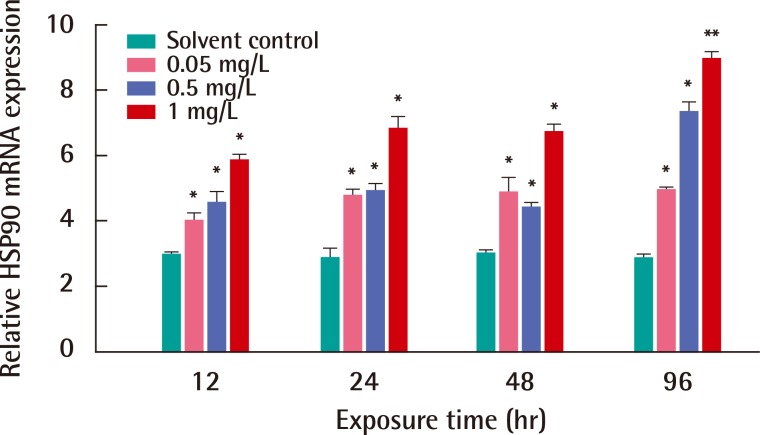
Expressions of Charybdis japonica heat shock protein 90 (HSP90) mRNA after exposure to various concentrations of bisphenol A for 12, 24, 48 and 96 hr. mRNA expression is shown relative to beta-actin expression after normalization. The experiment was conducted in triplicate and the data are expressed as mean±deviation. Differences between the treatment and solvent control samples were considered to be statistically significant when *p<0.05, **p<0.01.
Discussion
HSPs, also known as stress proteins and extrinsic chaperones, are a suite of highly conserved proteins with varying molecular weight produced in all cellular organisms when they are exposed to stress [32]. In the present study, an HSP90 gene from the Asian paddle crab C. japonica was identified. The deduced amino acid of C. japonica HSP90 shared a highly sequence similarity with other known HSP90s (over than 88%). The nucleotide sequence of C. japonica HSP90 shared a 96%, 91% and 89% identity with HSP90s sequences of Portunus trituberculatus, Scylla paramamosain and Chiromantes haematocheir crabs, respectively. In another study, C. japonica had a closer relationship with Callinectes sapidus and P. trituberculatus than with Scylla crabs using the complete mitochondrial DNA information [33]. The subfamily Portuninae has been included the genus Portunus, Charybdis and Scylla [33]. Given their economic importance and marvelous morphological diversity, the decapods have been studied most of all crustaceans [34]. However, we are far from reaching a consensus on their relationships among the decapods. Phylogenetic and network analysis suggest that the C. japonica HSP90 gene has an evolutionally close relationship with HSP90s of a lobster and with shrimps of other species (Figure 1). The gene information of crabs may be used for the monitoring in a marine ecosystem, although there is not enough molecular information about C. japonica crabs [28].
The real-time RT-PCR analysis revealed that C. japonica HSP90 gene is constitutively expressed with different levels in all reference tissues of crabs. In this study, the level of C. japonica HSP90 mRNA was the highest in ovary if compared with other tissues. This result was in agreement with the expression levels of HSP90s of marine crab (P. trituberculatus) [7], Chinese shrimp (Fenneropenaeus chinensis) [3] and black tiger shrimp ovary (Penaeus monodon) [6]. We deduced that C. japonica HSP90 might have a certain relationship with the genital organ development and maturation in an Asian paddle crab. However, expression of C. japonica HSP90 in hepatopancreas and gill as well as in ovary was also higher expressed than in other tissues, in the contrast to the ovary-specific expression of P. trituberculatus HSP90-1 gene [7].
HSP90, an abundant molecular chaperone, is an essential component of the protective stress response. HSP90s play key roles in signal transduction, cell-cycle control, genomic silencing and protein trafficking. It interacts with steroid hormone receptors, signaling kinases and various transcription factors. However, the mechanism by which HSP90 interacts with different proteins in various pathways remains unclear [35]. With exposure to EDCs treatments, an increase of C. japonica HSP90 mRNA was found in the Asian paddle crab exposed to various concentrations and exposure times. The significant expression of HSP90 mRNA was dose-dependent in C. japonica crab exposed to NP or BPA toxicity for 96 hours (Figures 3 and 4). Furthermore, the transcription of HSP90 was very sensitive in shrimp F. chinensis under environmental stress, such as heat shock and hypoxia [3]. The expression of HSP90 mRNA was induced in aquatic midge Chironomus riparius exposed to the EDC, such as di-2-ethylhexyl phthalate [32]. The response of HSP90 in atrazine treatments was significantly up-regulated in rare minnow (Grobiocypris rarus) [36].
EDCs mimic the action of endogenous estrogen hormones and interfere with the endocrine systems of a variety of organisms [19]. BPA and NP are well known EDCs ubiquitous in the aquatic environment and are an ecotoxicological risk for the health of aquatic organisms [20]. Distributions of these EDCs are widely found in the aquatic ecosystem, such as in rivers, lakes and coastal regions [37,38,39]. Potential genes have been used as sensitive biomarkers to assess EDCs toxicity in an aquatic environment. Choriogenin mRNA, regarded as sensitive biomarkers for estrogenic pollutants, is expressed in developing brackish medaka (Oryzias melastigma) after exposures of NP and BPA [40]. The expression of estrogen-related receptor genes was significantly up-regulated in C. riparius midge exposed to BPA and NP treated at all concentrations [19]. In the present study, we report significant responses of HSP90 gene in Asian paddle crab (C. japonica) after BPA and NP toxicity. Those results can be used as a highly sensitive biomarker for monitoring EDCs chemicals in the marine environment.
Acknowledgements
This study was supported by a grant of the National Research Foundation of Korea, which was funded by the Korean Government (NRF-2013R1A2A2A01004914).
Notes
The authors have no conflicts of interest with material presented in this paper.
This article is available from: http://e-eht.org/
